From Pixels to Insights: How AI and Vision Transformers Are Revolutionizing Bioimage Analysis

Advanced artificial intelligence (AI) is transforming biological research at unprecedented scales. Nowhere is this more evident than in bioimage analysis, the computational processing of images—from cells to entire tissues—to extract meaningful biological insights. AI-driven methods have automated complex tasks such as image restoration, cell segmentation, and phenotypic classification, which previously required extensive manual labor and domain expertise [1].
Central to this revolution have been convolutional neural networks (CNNs), a deep learning method that dominated bioimage processing over the past decade. CNNs excel at learning hierarchical image features and have delivered outstanding performance in tasks ranging from identifying cell types to detailed morphological profiling. Notable successes include tools like Cellpose, widely adopted for highly accurate cell segmentation in microscopy images [2].
In the recent years, a new generation of AI architecture—Vision Transformers (ViTs)—has emerged, inspired by transformer-based large language models (LLMs) such as GPT-3. ViTs treat image analysis differently, treating images as sequences of patches, analogous to words in a sentence. This strategy allows ViTs to capture long-range dependencies more effectively than CNNs, In additions ViTs scale more effectively than CNNs, becoming increasingly data-efficient and cost-effective as the dataset size grows [3].
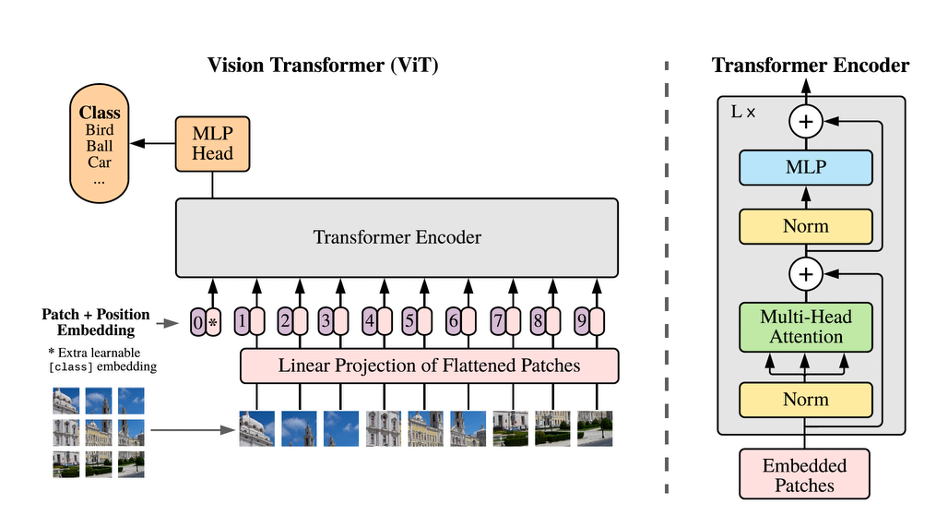
Fig. 1 An overview of the structure of a ViT model; Images are divided into patches and processed in a similar manner that a transformer-based LLM processes tokens in a sequence [1].
A prominent example is the self-supervised transformer model DINO (“DIstillation with NO labels”), which learns from unlabeled microscopy data. DINO achieved superior performance in classifying cell phenotypes by drug treatments compared to classical methods [4], while requiring no manual annotation. This advance illustrates ViTs’ potential for morphological profiling—rapidly and precisely grouping cells by their phenotypic responses to various drugs, crucial for accelerating drug discovery and understanding cellular mechanisms.
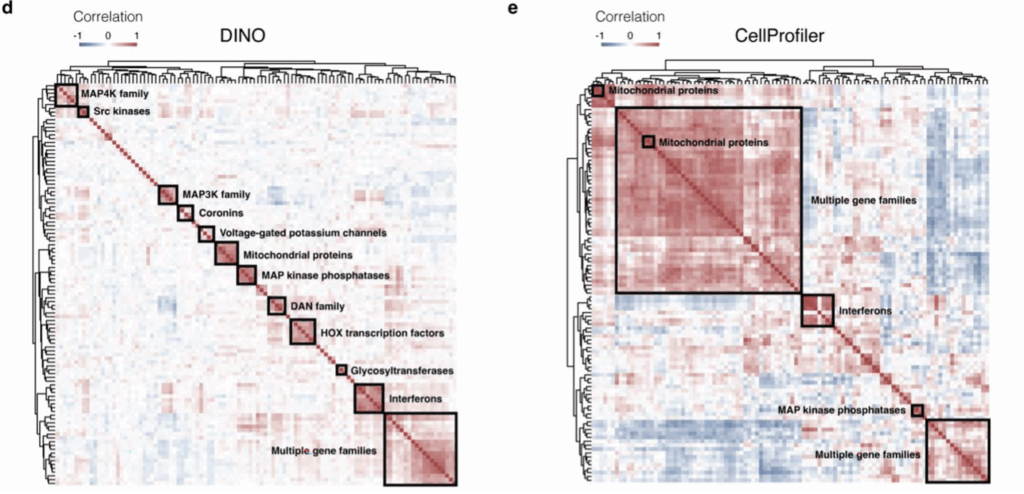
Figure. 2 Above compares the performance of DINO and Cellprofiler pipeline in grouping genes according to images of gene overexpression cell lines. The squares show groupings of genes that induce similar phenotypes. DINO recovers more detailed and higher quality groups [4].
In essence, ViTs represent a transformative step forward for bioimage analysis. By combining the strengths of transformers with the precision required for biological applications, ViTs promise a qualitative leap in how we derive biological insights from image data. For researchers and organizations focused on microscopy-based analyses, embracing these cutting-edge AI methods means unlocking deeper insights, faster and at larger scales than ever before.
References [1] Moen, E. et al. (2019). Deep learning for cellular image analysis. Nature Methods, 16(12), 1233–1246. [2] Stringer, C. et al. (2021). Cellpose: a generalist algorithm for cellular segmentation. Nature Methods, 18, 100–106. [3] Dosovitskiy, A. et al. (2021). An Image is Worth 16×16 Words: Transformers for Image Recognition at Scale. ICLR. [4] Tunyasuvunakool, K. et al. (2023). High-throughput morphological profiling using self-supervised vision transformers. Nature Biotechnology, 41(2), 181–190.
A great article by Milad Adibi !
